Runtime frontend
kmos.run
A front-end module to run a compiled kMC model. The actual model is
imported in kmc_model.so and all parameters are stored in kmc_settings.py.
The model can be used directly like so:
from kmos.model import KMC_Model
model = KMC_Model()
model.parameters.T = 500
model.do_steps(100000)
model.view()
which, of course can also be part of a python script.
The model can also be run in a different process using the
multiprocessing module. This mode is designed for use with
a GUI so that the CPU intensive kMC integration can run at
full throttle without impeding the front-end. Interaction with
the model happens through Queues.
-
class kmos.run.KMC_Model(image_queue=None, parameter_queue=None, signal_queue=None, size=None, system_name='kmc_model', banner=True, print_rates=True, autosend=True)
API Front-end to initialize and run a kMC model using python bindings.
Depending on the constructor cal the model can be run either via directory
calls or in a separate processes access via multiprocessing.Queues.
Only one model instance can exist simultaneously per process.
-
deallocate()
Deallocate all arrays that are allocated
by the Fortran module. This needs to be called
whenever more than one simulation is started
from one process.
Note that the currenty state and history of
the system is lost after calling this method.
Note: explicit invocation was chosen over the
__del__ method because there seems to easy
portable way to control garbage collection.
-
do_steps(n=10000)
Propagate the model n steps.
-
double()
Double the size of the model in each direction and initialize
larger model with current configuration in each copy.
-
get_atoms(geometry=True)
Return an ASE Atoms object with additional
information such as coverage and Turn-over-frequencies
attached.
Returns the names of the fields returned by
self.get_atoms().occupation.
Useful for the header line of an ASCII output.
Returns the names of the fields returned by
self.get_atoms().tof_data.
Useful for the header line of an ASCII output.
-
halve()
Halve the size of the model and initialize each site in the new model
with a species randomly drawn from the sites that are reduced onto
one. It is necessary that the simulation size is even.
-
post_mortem(steps=None, propagate=False, err_code=None)
Accepts an integer and generates a post-mortem report
by running that many steps and returning which process
would be executed next without executing it.
-
put(site, new_species)
Puts new_species at site. The site is given by 4-entry sequence
like [x, y, z, n], where the first 3 entries define the unit cell
from 0 to the number of unit cells in the respective direction.
And n specifies the site within the unit cell.
The database of available processes will be updated automatically.
Examples
model.put([0,0,0,model.lattice.lattice_bridge], model.proclist.co ])
# puts a CO molecule at the `bridge` site of the lower left unit cell
-
_put(site, new_species)
Works exactly like put, but without updating the database of
available processes. This is faster for when one does a lot updates
at once, however one must call _adjust_database afterwards.
Examples
model._put([0,0,0,model.lattice.lattice_bridge], model.proclist.co ])
# puts a CO molecule at the `bridge` site of the lower left unit cell
model._put([1,0,0,model.lattice.lattice_bridge], model.proclist.co ])
# puts a CO molecule at the `bridge` site one to the right
# ... many more
model._adjust_database() # Important !
-
run()
Runs the model indefinitely. To control the
simulations, model must have been initialized
with proper Queues.
-
show_accum_rate_summation(order='-rate')
Shows rate individual processes contribute to the total rate
The optional argument order can be one of: name, rate, rate_constant,
nrofsites. You precede each keyword with a ‘-‘, to show in decreasing
order.
Default: ‘-rate’.
-
start()
Start child process
-
view()
Start current model in live view mode.
-
xml()
Returns the XML representation that this model was created from.
-
_get_configuration()
Return current configuration of model.
-
_set_configuration(config)
Set the current lattice configuration.
Expects a 4-dimensional array, with dimensions [X, Y, Z, N]
where X, Y, Z are the lattice size and N the number of
sites in each unit cell.
-
class kmos.run.Model_Rate_Constants
Holds all rate constants currently associated with the model.
To inspect the expression and current settings of it you can just
call it as a function with a (glob) pattern that matches
the desired processes, e.g.
model.rate_constant('*ads*')
could print all rate constants for adsorption. Given of course that
‘ads’ is part of the process name. The just get the rate constant
for one specific process you can use
model.rate_constant.by_name("<process name>")
To set rate constants manually use
model.rate_constants.set("<pattern>", <rate-constant (expr.)>)
-
class kmos.run.Model_Parameters(print_rates=True)
Holds all user defined parameters of a model in
concise form. All user defined parameters can be
accessed and set as attributes, like so
model.parameters.<parameter> = X.Y
kmos.view
Run and view a kMC model. For this to work one needs a
kmc_model.(so/pyd) and a kmc_settings.py in the import path.
-
class kmos.view.KMC_Viewer(model=None)
A graphical front-end to run, manipulate
and view a kMC model.
-
exit(_widget, _event)
Exit the viewer application cleanly
killing all subprocesses before the main
process.
-
parameter_callback(name, value)
Sent (updated) parameters to the model process.
kmos.utils
Several utility functions that do not seem to fit somewhere
else.
-
kmos.utils.build(options)
Build binary with f2py binding from complete
set of source file in the current directory.
-
kmos.utils.evaluate_kind_values(infile, outfile)
Go through a given file and dynamically
replace all selected_int/real_kind calls
with the dynamically evaluated fortran code
using only code that the function itself
contains.
-
kmos.utils.get_ase_constructor(atoms)
Return the ASE constructor string for atoms.
-
kmos.utils.split_sequence(seq, size)
Take a list and a number n and return list
divided into n sublists of roughly equal size.
-
kmos.utils.write_py(fileobj, images, **kwargs)
Write a ASE atoms construction string for images
into fileobj.
kmos kMC project DTD
The central storage and exchange format is XML. XML was chosen over
JSON, pickle or alike because it still seems as the most flexible
and universal format with good methods to define the overall
structure of the data.
One way to define an XML format is by using a document type description
(DTD) and in fact at every import a kmos file is validated against
the DTD below.
<!ELEMENT kmc (meta?,species_list?,parameter_list?, lattice, process_list?,output_list?)>
<!ATTLIST kmc
version CDATA #REQUIRED
>
<!ELEMENT meta EMPTY>
<!ATTLIST meta
author CDATA #IMPLIED
debug CDATA #IMPLIED
email CDATA #IMPLIED
model_dimension CDATA #IMPLIED
model_name CDATA #IMPLIED
>
<!ELEMENT species_list (species)*>
<!ATTLIST species_list
default_species CDATA #IMPLIED
>
<!ELEMENT species EMPTY>
<!ATTLIST species
name CDATA #REQUIRED
color CDATA #IMPLIED
representation CDATA #IMPLIED
tags CDATA #IMPLIED
>
<!ELEMENT parameter_list (parameter)*>
<!ELEMENT parameter EMPTY>
<!ATTLIST parameter
name CDATA #REQUIRED
value CDATA #IMPLIED
adjustable CDATA #IMPLIED
min CDATA #IMPLIED
max CDATA #IMPLIED
scale CDATA #IMPLIED
>
<!ELEMENT lattice (layer)*>
<!ATTLIST lattice
cell_size CDATA #REQUIRED
default_layer CDATA #REQUIRED
substrate_layer CDATA #IMPLIED
representation CDATA #IMPLIED
>
<!ELEMENT layer (site)*>
<!ATTLIST layer
name CDATA #REQUIRED
grid CDATA #IMPLIED
grid_offset CDATA #IMPLIED
color CDATA #IMPLIED
>
<!ELEMENT site EMPTY>
<!ATTLIST site
pos CDATA #REQUIRED
type CDATA #REQUIRED
tags CDATA #IMPLIED
default_species CDATA #IMPLIED
>
<!ELEMENT process_list (process)*>
<!ELEMENT process (condition|action)*>
<!ATTLIST process
name CDATA #REQUIRED
rate_constant CDATA #REQUIRED
enabled CDATA #IMPLIED
tof_count CDATA #IMPLIED
>
<!ELEMENT condition EMPTY>
<!ATTLIST condition
coord_name CDATA #REQUIRED
coord_layer CDATA #REQUIRED
coord_offset CDATA #REQUIRED
species CDATA #REQUIRED
>
<!ELEMENT action EMPTY>
<!ATTLIST action
coord_name CDATA #REQUIRED
coord_layer CDATA #REQUIRED
coord_offset CDATA #REQUIRED
species CDATA #REQUIRED
>
<!ELEMENT output_list (output)*>
<!ELEMENT output EMPTY>
<!ATTLIST output
item CDATA #REQUIRED
>
Backend
In general the backend includes all functions that are implemented in Fortran90,
which therefore should not have to be changed by hand often. The backend is
divided into three modules, which import each other in the following way
base <- lattice <- proclist
The key for this division is reusability of the code. The base module
implement all aspects of the kMC code, which do not depend on the described
model. Thus it “never” has to change. The latttice module basically
repeats all methods of the base model in terms of lattice coordinates.
Thus the lattice module only changes, when the geometry of the model
changes, e.g. when you add or delete sites.
The proclist module implements the process list, that is the species
or states each site can have and the elementary steps. Typically that
changes most often while developing a model.
The rate constants and physical parameters of the system are not implemented
in the backend at all, since in the physical sense they are too high-level
to justify encoding and compilation at the Fortran level and so they
are typical read and parsed from a python script.
The kmos.run.KMC_Model class implements a convenient interface for most of
these functions, however all public methods (in Fortran called subroutines)
and variables can also be accessed directly like so
from kmos.run import KMC_Model
model = KMC_Model(print_rates=False, banner=False)
model.base.<TAB>
model.lattice.<TAB>
model.proclist.<TAB>
which works best in conjunction with ipython.
kmos/base
The base kMC module, which implements the kMC method on a 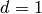 lattice. Virtually any lattice kMC model can be build on top of this.
The methods offered are:
lattice. Virtually any lattice kMC model can be build on top of this.
The methods offered are:
- de/allocation of memory
- book-keeping of the lattice configuration and all available processes
- updating and tracking kMC time, kMC step and wall time
- saving and reloading the current state
- determine the process and site to be executed
base/accum_rates
Stores the accumulated rate constant multiplied with the number
of sites available for that process to be used by determine_procsite.
Let

be the rate constants
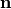
the number of available sites, and
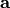
the accumulated rates, then
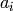
is calculated according to
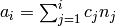
.
base/add_proc
The main idea of this subroutine is described in del_proc. Adding one
process to one capability is programmatically simpler since we can just
add it to the end of the respective array in avail_sites.
- proc positive integer number that represents the process to be added.
- site positive integer number that represents the site to be manipulated
base/allocate_system
Allocates all book-keeping structures and stores
local copies of system name and size(s):
- systen_name identifier of this simulation, used as name of punch file
- volume the total number of sites
- nr_of_proc the total number of processes
base/assertion_fail
Function that shall be used by all parts of the program to print a
proper message in case some assertion fails.
- a condition that is supposed to hold true
- r message that is printed to the poor user in case it fails
base/avail_sites
Main book-keeping array that stores for each process the sites
that are available and for each site the address
in this very array. The meaning of the fields are:
avail_sites(proc, field, switch)
where:
- proc – refers to a process in the process list
- the field within the process, but the meaning differs as explained
under ‘switch’
- switch – can be either 1 or 2 and switches between
(1) the actual numbers of the sites, which are available
and filled in from the left but in whatever order they come
or (2) the location where the site is stored in (1).
base/can_do
Returns true if ‘site’ can do ‘proc’ right now
- proc integer representing the requested process.
- site integer representing the requested site.
- can writeable boolean, where the result will be stored.
base/deallocate_system
Deallocate all allocatable arrays: avail_sites, lattice, rates,
accum_rates, procstat.
none
base/del_proc
del_proc delete one process from the main book-keeping array
avail_sites. These book-keeping operations happen in O(1) time with the
help of some more book-keeping overhead. avail_sites stores for each
process all sites that are available. The array for each process is
filled from the left, but sites generally not ordered. With this
determine_procsite can effectively pick the next site and process. On
the other hand a second array (avail_sites(:,:,2) ) holds for each
process and each site, the location where it is stored in
avail_site(:,:,1). If a site needs to be removed this subroutine first
looks up the location via avail_sites(:,:,1) and replaces it with the
site that is stored as the last element for this process.
- proc positive integer that states the process
- site positive integer that encodes the site to be manipulated
base/get_accum_rate
Return accumulated rate at a given process.
- proc_nr integer representing the requested process.
- return_accum_rate writeable real, where the requested accumulated rate will be stored.
base/get_avail_site
Return field from the avail_sites database
- proc_nr integer representing the requested process.
- field integer for the site at question
- switch 1 or 2 for site or storage location
base/get_kmc_step
Return the current kmc_step
- kmc_step Writeable integer
base/get_kmc_time
Returns current kmc_time as rdouble real as defined in kind_values.f90.
- return_kmc_time writeable real, where the kmc_time will be stored.
base/get_kmc_time_step
Returns current kmc_time_step (the time increment).
- return_kmc_step writeable real, where the kmc_time_step will be stored.
base/get_kmc_volume
Return the total number of sites.
- volume Writeable integer.
base/get_nrofsites
Return how many sites are available for a certain process.
Usually used for debugging
- proc integer representing the requested process
- return_nrofsites writeable integer, where nr of sites gets stored
base/get_procstat
Return process counter for process proc as integer.
- proc integer representing the requested process.
- return_procstat writeable integer, where the process counter will be stored.
base/get_rate
Return rate of given process.
- proc_nr integer representing the requested process.
- return_rate writeable real, where the requested rate will be stored.
base/get_species
Return the species that occupies site.
- site integer representing the site
base/get_system_name
Return the systems name, that was specified with base/allocate_system
- system_name Writeable string of type character(len=200).
base/get_walltime
Return the current walltime.
- return_walltime writeable real where the walltime will be stored.
base/increment_procstat
Increment the process counter for process proc by one.
- proc integer representing the process to be increment.
base/interval_search_real
This is basically a standard binary search algorithm that expects an array
of ascending real numbers and a scalar real and return the key of the
corresponding field, with the following modification :
- the value of the returned field is equal of larger of the given
value. This is important because the given value is between 0 and the
largest value in the array and otherwise the last field is never
selected.
- if two or more values in the array are identical, the function
return the index of the leftmost of those field. This is important
because having field with identical values means that all field except
the leftmost one do not contain any sites. Refer to
update_accum_rate to understand why.
- the value of the returned field may no be zero. Therefore the index
the to be equal or larger than the first non-zero field.
However: as everyone knows the binary search is trickier than it appears
at first site especially real numbers. So intensive testing is
suggested here!
- arr real array of type rsingle (kind_values.f90) in monotonically (not strictly) increasing order
- value real positive number from [0, max_arr_value]
base/kmc_step
Number of kMC steps executed.
base/kmc_time
Simulated kMC time in this run in seconds.
base/kmc_time_step
The time increment of the current kMC step.
base/lattice
Stores the actual physical lattice in a 1d array, where the value
on each slot represents the species on that site.
Species constants can be conveniently defined
in lattice_... and later used directly in the process list.
base/nr_of_proc
Total number of available processes.
base/nr_of_sites
Stores the number of sites available for each process.
base/procstat
Stores the total number of times each process has been executed
during one simulation.
base/rates
Stores the rate constants for each process in s^-1.
base/reload_system
Restore state of simulation from *.reload file as saved by
save_system(). This function also allocates the system’s memory
so calling allocate_system again, will cause a runtime failure.
- system_name string of 200 characters which will make the reload_system look for a file called ./<system_name>.reload
- reloaded logical return variable, that is .true. reload of system could be completed successfully, and .false. otherwise.
base/replace_species
Replaces the species at a given site with new_species, given
that old_species is correct, i.e. identical to the site that
is already there.
- site integer representing the site
- old_species integer representing the species to be removed
- new_species integer representing the species to be placed
base/reset_site
This function is a higher-level function to reset a site
as if it never existed. To achieve this the species
is set to null_species and all available processes
are stripped from the site via del_proc.
- site integer representing the requested site.
- species integer representing the species that ought to be at the site, for consistency checks
base/save_system
save_system stores the entire system information in a simple ASCII
filed names <system_name>.reload. All fields except avail_sites are
stored in the simple scheme:
variable value
In the case of array variables, multiple values are seperated by one or
more spaces, and the record is terminated with a newline. The variable
avail_sites is treated slightly differently, since printed on a single
line it is almost impossible to interpret from the ASCII files. Instead
each process starts a new line, and the first number on the line stands
for the process number and the remaining fields, hold the values.
none
base/set_kmc_time
Sets current kmc_time as rdouble real as defined in kind_values.f90.
- new readable real, that the kmc time will be set to
base/set_rate_const
Allows to set the rate constant of the process with the number proc_nr.
- proc_n The process number as defined in the corresponding proclist_ module.
- rate the rate in
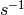
base/set_system_name
Set the systems name. Useful in conjunction with base.save_system
to save *.reload files under a different name than the default one.
- system_name Readable string of type character(len=200).
base/start_time
CPU time spent in simulation at least reload.
base/system_name
Unique indentifier of this simulation to be used for restart files.
This name should not contain any characters that you don’t want to
have in a filename either, i.e. only [A-Za-z0-9_-].
base/update_accum_rate
Updates the vector of accum_rates.
none
base/update_clocks
Updates walltime, kmc_step and kmc_time.
- ran_time Random real number
![\in [0,1]](../_images/math/5a8fb07940437396dd240303860b93950e70ce3d.png)
base/volume
Total number of sites.
base/walltime
Total CPU time spent on this simulation.
kmos/lattice
Implements the mappings between the real space lattice
and the 1-D lattice, which kmos/base operates on.
Furthermore replicates all geometry specific functions of kmos/base
in terms of lattice coordinates.
Using this module each site can be addressed with 4-tuple
(i, j, k, n) where i, j, k define the unit cell and
n the site within the unit cell.
lattice/allocate_system
Allocates system, fills mapping cache, and
checks whether mapping is consistent
none
lattice/calculate_lattice2nr
Maps all lattice coordinates onto a continuous
set of integer ![\in [1,volume]](../_images/math/be7459e2d0b8d4daf6242fc283f26a20d9d23c75.png)
- site integer array of size (4) a lattice coordinate
lattice/calculate_nr2lattice
Maps a continuous set of
of integers ![\in [1,volume]](../_images/math/be7459e2d0b8d4daf6242fc283f26a20d9d23c75.png) to a
4-tuple representing a lattice coordinate
to a
4-tuple representing a lattice coordinate
- nr integer representing the site index
lattice/deallocate_system
Deallocates system including mapping cache.
none
kmos/proclist
Implements the kMC process list.
proclist/do_kmc_step
Performs exactly one kMC step.
none
proclist/get_kmc_step
Determines next step without executing it.
none
proclist/get_occupation
Evaluate current lattice configuration and returns
the normalized occupation as matrix. Different species
run along the first axis and different sites run
along the second.
none
proclist/init
Allocates the system and initializes all sites in the given
layer.
- input_system_size number of unit cell per axis.
- system_name identifier for reload file.
- layer initial layer.
- no_banner [optional] if True no copyright is issued.
proclist/initialize_state
Initialize all sites and book-keeping array
for the given layer.
- layer integer representing layer
proclist/run_proc_nr
Runs process proc on site nr_site.
- proc integer representing the process number
- nr_site integer representing the site

 lattice. Virtually any lattice kMC model can be build on top of this.
The methods offered are:
lattice. Virtually any lattice kMC model can be build on top of this.
The methods offered are: be the rate constants
be the rate constants 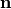 the number of available sites, and
the number of available sites, and 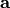 the accumulated rates, then
the accumulated rates, then 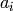 is calculated according to
is calculated according to 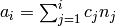 .
.![\in[0,1]](../_images/math/ad1f4f4523e3a6994e94149f5cdbf296a9792988.png) that selects the next process
that selects the next process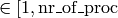
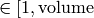
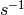
![\in [0,1]](../_images/math/5a8fb07940437396dd240303860b93950e70ce3d.png)
![\in [1,volume]](../_images/math/be7459e2d0b8d4daf6242fc283f26a20d9d23c75.png)